Next: GA with Genotype and Up: Example: 'Goldberg Variations' Previous: Population: P, P' Contents
Accordingly we need a fitness function
![]() with the above mentioned properties. Goldberg proposed the simple function
with the above mentioned properties. Goldberg proposed the simple function
| (5.11) | |||
| (5.12) | |||
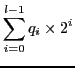 |
(5.13) |
Using our population ![]() from above we can identify the first two elements and translate these binary patterns into decimal numbers with the aid of the scilab function
from above we can identify the first two elements and translate these binary patterns into decimal numbers with the aid of the scilab function ![]() from program GA_v4.sce:
from program GA_v4.sce:
-->element1=Pwork(1,:)
element1 =
0. 1. 0. 0. 0.
-->l=5,[d]= vec2dec(element1,l)
l =
5.
d =
8.
-->element2=Pwork(2,:)
element2 =
0. 1. 0. 0. 1.
-->l=5,[d]= vec2dec(element2,l)
l =
5.
d =
9.
The fitness value ![]() can then easily be computed. With the possible members in the realm of
can then easily be computed. With the possible members in the realm of
![]() we have a possible range of fitness values between
we have a possible range of fitness values between
![]() . If we assume as a possible threshold
. If we assume as a possible threshold
![]() then we have as set of selected fitness values
then we have as set of selected fitness values ![]() . Then we get as maximal value
. Then we get as maximal value
![]() . Then we get as possible goal for a genetic process
. Then we get as possible goal for a genetic process
![]() . Thus we will have '124' as '100%'.
. Thus we will have '124' as '100%'.
One simulation of this population ![]() could then show results (cf. diagram 5.11) where the fitness of the whole population stays below 30% all the time when not using mutation.
could then show results (cf. diagram 5.11) where the fitness of the whole population stays below 30% all the time when not using mutation.
Invoking a simulation using the scilab function
![]() from the program GA_v4.sce:
from the program GA_v4.sce:
-->POP=Pwork, l=5,p=5,n=4,run=100,MThreshold=run,[POP,FITNESS_ALLLOG]=gasimple(POP,l,p,n,run, MThreshold)
Gerd Doeben-Henisch 2013-01-14